Yubexi Correa1, Rita Del Giudice1, Sarah Waldie1,2a,3, Michel Thépaut4, Samantha Micciula2b, Yuri Gerelli5,6, Martine Moulin2a,3, Clara Delaunay4, Franck Fieschi3,4,7, Harald Pichler8, Michael Haertlein2a,3, V. Trevor Forsyth2a,3,9, Anton Le Brun10, Michael Moir10, Robert A. Russell10, Tamim Darwish10, Jonas Brinck11, Tigist Wodaje11, Martin Jansen12, César Martín13, Felix Roosen-Runge1 and Marité Cárdenas1,13,14,15.
1 Biofilm – Research Center for Biointerfaces and Department of Biomedical Science, Faculty of Health and Society, Malmö University, 20506 Malmö, Sweden.
2 aLife Sciences Group, Institut Laue Langevin, Grenoble F-38042, France. bLarge Scale Structures, Institut Laue Langevin (ILL), Grenoble F-38042, France.
3 Partnership for Structural Biology, Grenoble F-38042, France
4 Univ. Grenoble Alpes, CNRS, CEA, IBS, 71 avenue des Martyrs, F-38000 Grenoble, France.
5 Marche Polytechnic University, Department of Life and Environmental Sciences, Via Brecce Bianche 12, 60131, Ancona, Italy
6 CNR-ISC and Department of Physics, Sapienza University of Rome, Piazzale A. Moro 2, Rome, Italy
7 Institut universitaire de France (IUF), Paris, France.
8 1aAustrian Centre of Industrial Biotechnology, Petersgasse 14, 8010, Graz, Austria. b Graz University of Technology, Institute of Molecular Biotechnology, NAWI Graz, BioTechMed Graz, Petersgasse 14, 8010, Graz, Austria.
9 aFaculty of Medicine, Lund University, 22184 Lund, Sweden. bLINXS Institute for Advanced Neutron and X-ray Science, Scheelevagen 19, 22370 Lund.
10 National Deuteration Facility, Australian Nuclear Science and Technology Organization (ANSTO), New Illawarra Road, Lucas Heights, NSW 2234, Australia.
11 Karolinska Institute, Stockholm, Sweden
12 Institute of Clinical Chemistry and Laboratory Medicine, Medical Centre, University of Freiburg, Freiburg Im Breisgau, Germany
13 Department of Molecular Biophysics, Biofisika Institute (University of Basque Country and Consejo Superior de Investigaciones Científicas (UPV/EHU, CSIC)), 48940 Leioa, Spain
14 School of Biological Sciences, Nanyang Technological University, Singapore
15 IKERBASQUE, Basque Foundation for Science, Bilbao, Spain
See the full published paper: Correa. et al., Journal of Colloid and Interface Science, 645 (2023) 627-638.
Since the outbreak of the COVID-19 pandemics at the end of December 2019, much has been discussed about the relationship between the severity of the disease and the plasma lipid profiles of the population1. For this reason, we were curious about the role that plays the Spike protein (S protein) from SARS-CoV-2 in the lipid metabolisms, especially its interaction with High-Density Lipoproteins (HDLs), one of the main groups of lipoproteins that transport cholesterol from peripheral tissue to the liver where it is recycled or removed from the body2.
This led us to our first work on the topic3, on which we showed that the S protein removes lipids from model membranes and interferes with the capacity of HDL for removing and exchanging lipids composed of saturated phospholipids (DMPC) and cholesterol. This work left behind open questions such as: does the S protein have preference over certain lipids? Could the lipid serum profile in individuals have an effect over how the S protein affect HDL function?
In our subsequent paper, we tested our hypothesis by using various types of lipids (saturated, monounsaturated and polyunsaturated). We took advantage of deuteration and neutron reflection to determine the various affinities for either HDL or the S protein for such lipids. Due to the differences in scattering length density between hydrogen and deuterium, the lipid removal and deposition by HDL (characteristic to its function) can be resolved in detail (Figure 1.I). We found that the S protein had the strongest preference for saturated lipids in the presence of cholesterol and polyunsaturated lipids containing linoleic acid (Figure 1.II).

We then focused on how the actual plasma composition in terms of total triglycerides (TG) and total cholesterol (TC) regulated the ability of the S protein to affect HDL function. Our data suggest that individuals with a particular plasma lipid profile (low triglyceride and high cholesterol) may be less likely to have their lipid metabolism hijacked during COVID-19 infection or following COVID-19 immunization since there is a weaker interaction with the S protein with HDL in this case (Figure 2).
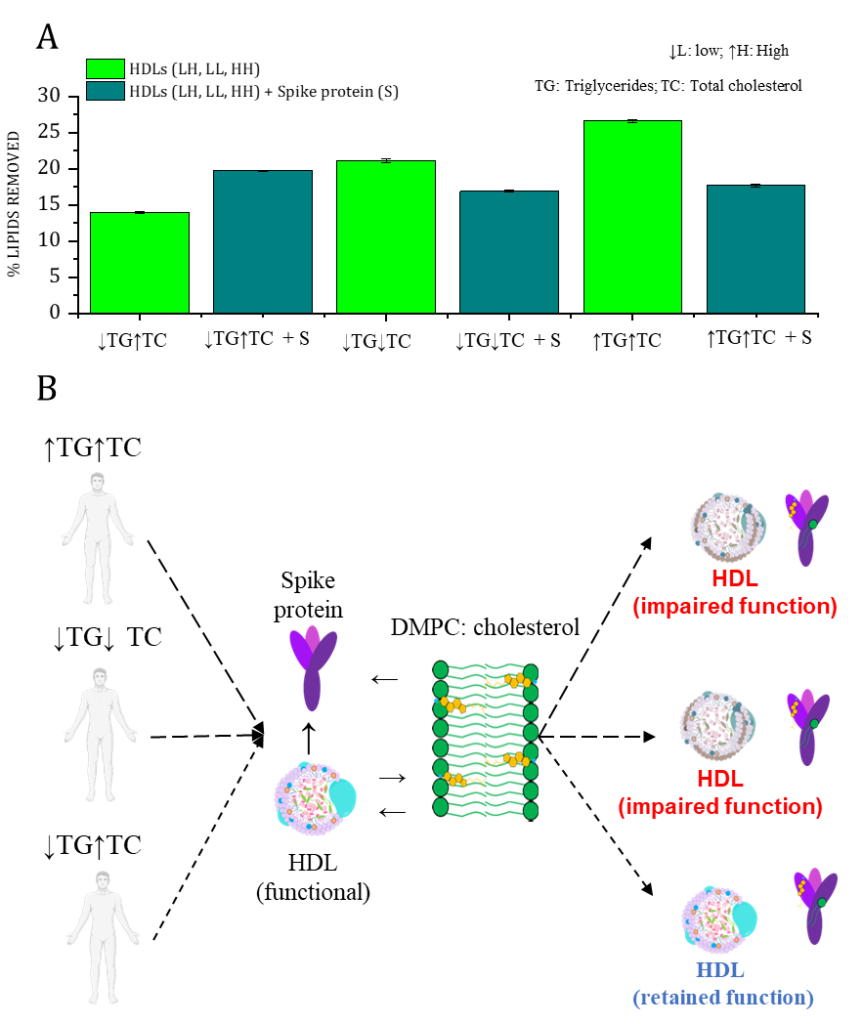
Figure 2. A) Effect of the S protein on HDL capacity to remove lipids on HDL samples from individuals with low triglycerides/high cholesterol (↓TG↑TC), low triglycerides/low cholesterol (↓TG↓TC), and high triglycerides/high cholesterol (↑TG↑TC) plasma levels. Data was obtained from neutron reflection profiles after 5 hours of incubation of the SLB (d-DMPC: d-cholesterol, 80:20 mol%) with either HDL (0.132 mg/mL), or a mixture of both HDL and S protein (0.132 and 0.05 mg/mL, respectively) in h-TBS at 37 °C. B) Schematic representation of the interaction between HDL and the S protein as a function of the lipid profile of individuals. Spike protein modulates HDL function depending on the donor lipid profile.
These experiments were performed thanks to the Collaboration with the deuteration labs at Institut Laue-Langevin (ILL) and Australian Centre for Neutron Scattering (ANSTO), as well as to beamtime granted at ILL on Figaro and D17, as well as at ANSTO on Spatz neutron reflectometers. Importantly, the neutron reflection data mirrored the traditional biochemical assay for reverse cholesterol transport of HDL, on which the transfer of cholesterol from macrophages to HDL is followed.
References
[1] Mahat RK, Rathore V, Singh N, Singh N, Singh SK, Shah RK, Garg C. Lipid profile as an indicator of COVID-19 severity: A systematic review and meta-analysis. Clin Nutr ESPEN. 2021 Oct; 45:91-101. doi: 10.1016/j.clnesp.2021.07.023. Epub 2021 Jul 31. PMID: 34620375; PMCID: PMC8325550.
[2] M.P. Adorni, N. Ronda, F. Bernini, F. Zimetti, High density lipoprotein cholesterol efflux capacity and atherosclerosis in cardiovascular disease: Pathophysiological aspects and pharmacological perspectives, Cells. 10 (2021) 1–37. https://doi.org/10.3390/cells10030574.
[3] Y. Correa, S. Waldie, M. Thépaut, S. Micciula, M. Moulin, F. Fieschi, H. Pichler, V. Trevor Forsyth, M. Haertlein, M. Cárdenas. SARS-CoV-2 spike protein removes lipids from model membranes and interferes with the capacity of high-density lipoprotein to exchange lipids. J. Colloid Interface Sci., 602 (2021), pp. 732-739, 10.1016/j.jcis.2021.06.056







